Utilisateur:Amarchais/RsaOG RNA
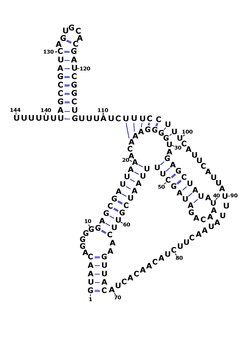
RsaOG (acronym for RNA S. aureus Orsay G) is a non-coding RNA that was discovered in the pathogenic bacteria Staphylococcus aureus N315 using a large scale computational screening based on phylogenetic profiling[1]. It was subsequently identified in other strains under the name of RsaI[2]. The rsaOG gene is conserved in all Staphylococcaceae sequenced genomes. Northern blot experiments show that RsaOG is expressed in several S. aureus strains[1],[2]. Mapping of RsaOG ends indicates a size of 146 nucleotides in S. aureus[2]. The rsaOG function is unknown.
References[modifier | modifier le code]
- Geissmann T, Chevalier C, Cros MJ, et al., « A search for small noncoding RNAs in Staphylococcus aureus reveals a conserved sequence motif for regulation », Nucleic Acids Res., (PMID 19786493, DOI 10.1093/nar/gkp668)
External links[modifier | modifier le code]
[[Category:Non-coding RNA]] {{molecular-cell-biology-stub}} RsaOG Alignment [1]
