Abstract
Hutchinson–Gilford progeria syndrome (HGPS) is a rare lethal genetic disorder characterized by symptoms reminiscent of accelerated aging. The major underlying genetic cause is a substitution mutation in the gene coding for lamin A, causing the production of a toxic isoform called progerin. Here we show that reduction of lamin A/progerin by a single-dose systemic administration of adeno-associated virus-delivered CRISPR–Cas9 components suppresses HGPS in a mouse model.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Data availability
The accession number for the RNA-Seq data reported in this paper is NCBI GEO: GSE122865.
References
Worman, H. J., Fong, L. G., Muchir, A. & Young, S. G. J. Clin. Invest. 119, 1825–1836 (2009).
Broers, J. L., Ramaekers, F. C., Bonne, G., Yaou, R. B. & Hutchison, C. J. Physiol. Rev. 86, 967–1008 (2006).
Kubben, N. & Misteli, T. Nat. Rev. Mol. Cell Biol. 18, 595–609 (2017).
Gordon, L. B., Rothman, F. G., Lopez-Otin, C. & Misteli, T. Cell 156, 400–407 (2014).
Ahmed, M. S., Ikram, S., Bibi, N. & Mir, A. Mol. Neurobiol. 55, 4417–4427 (2018).
Gonzalo, S., Kreienkamp, R. & Askjaer, P. Ageing Res. Rev. 33, 18–29 (2017).
Gordon, L. B. et al. Circulation 130, 27–34 (2014).
Gordon, L. B. et al. GeneReviews® [Internet] (NCBI Bookshelf, 2003).
Young, S. G., Yang, S. H., Davies, B. S., Jung, H. J. & Fong, L. G. Sci. Transl. Med. 5, 171ps3 (2013).
Vidak, S. & Foisner, R. Histochem. Cell. Biol. 145, 401–417 (2016).
De Sandre-Giovannoli, A. et al. Science 300, 2055 (2003).
Eriksson, M. et al. Nature 423, 293–298 (2003).
Buchwalter, A. & Hetzer, M. W. Nat. Commun. 8, 328 (2017).
Osorio, F. G. et al. Sci. Transl. Med. 3, 106ra107 (2011).
Fong, L. G. et al. J. Clin. Invest. 116, 743–752 (2006).
Lopez-Mejia, I. C. et al. EMBO Rep. 15, 529–539 (2014).
Olaya Santiago-Fernández, F. G. O. et al. Nat. Med. (in the press).
Ocampo, A. et al. Cell 167, 1719–1733 e12 (2016).
Yang, S. H. et al. Hum. Mol. Genet. 24, 2826–2840 (2015).
Liao, H. K. et al. Cell 171, 1495–1507 e15 (2017).
Dobin, A. et al. Bioinformatics 29, 15–21 (2013).
Acknowledgements
We thank A. Ocampo for discussions throughout the study and M. Schwarz for administrative support. We are grateful to G. Sancar and Ronald Evans Lab for providing the running wheel equipment. E.B. was partially funded by the Catharina Foundation. P.R. was partially supported by the Muscular Dystrophy Association. G.E. was partially funded by NIH-NCI CCSG: P30 014195 and by the Helmsley Trust. Work in the laboratory of J.C.I.B. was supported by The Progeria Research Foundation, Universidad Católica San Antonio de Murcia (UCAM), Fundacion Dr. Pedro Guillen, the G. Harold and Leila Y. Mathers Charitable Foundation, The Glenn Foundation and The Moxie Foundation.
Author information
Authors and Affiliations
Contributions
E.B., H.-K.L. and J.C.I.B. designed all the experiments. E.B., H.-K.L., P.R. and J.C.I.B. prepared the figures and wrote the manuscript. E.B., H.-K.L. and Y.F. performed and/or analyzed in vitro experiments. E.B., H.-K.L., Y.F., M.Y., R.H.-B. and P.R. performed and/or analyzed ex vivo and in vivo experiments. G.E. performed the bioinformatics analyses.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
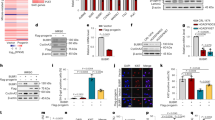
Extended Data Fig. 1 Immunoblots and RNA sequencing of Lmna gRNA-treated fibroblasts of adult HGPS mice.
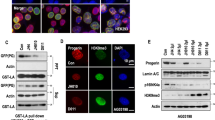
a, Immunoblots of individual (g1 or g2) or multiplex (g1&2) gRNA-treated fibroblasts derived from adult HGPS (Pro); Cas9 mice. Fibroblasts derived from heterozygous (Pro/+) and homozygous (Pro/Pro) HGPS mice of a heterozygous Cas9 (Cas9/+) background were treated with lentivirally delivered gRNAs. No Cas9 (Pro/+;+/+) sample or mock-treatments were used as negative controls. Middle panels show longer exposures. α-Tubulin and β-actin were used as loading controls. b, Relative intensities of lamin A, progerin and lamin C bands on the immunoblots. The intensity of each band was normalized by β-actin intensity and compared to normalized mock-control. c, Custom tracks of RNA sequences identified with high-throughput sequencing. The magnified region corresponds to the square blue area. The red line denotes the truncated region in HGPS (Pro). Shown are RNA samples derived from mouse fibroblasts of the indicated genotypes and treatments: + denotes WT. Mock-treatments and heterozygous HGPS mice without Cas9 (Pro/+; +/+) were used as negative controls. The arrows are not drawn to scale. d, Quantification of RNA sequencing analyses shows reduced transcription of exons 11 and 12 in Cas9-positive cells treated with gRNAs relative to mock-treated or Cas9-negative cells.
Extended Data Fig. 2 Fluorescent images of Lmna gRNA-injected mice.
a, Whole-body fluorescent images of a P4 (4 days dpp, days postpartum) mouse injected at P0 with a 1:1 mix of gRNA1 and gRNA2 viral preparations (each 1.29 × 1011 genomic copies per viral preparation). b, Same organs as in Fig. 1c, under individually adjusted exposure levels relative to negative controls. The organs were harvested from a 6 dpp mouse, 5 days post-injection (dpi) at P1 (P1-5DPI). The inset corresponds to the square area. Ctrl: no injection. c, Expression of the mCherry reporter in the adult (5.5 months post-injection of a 1:1 mix of gRNA1 and 2 viral preparations;1.3 × 1011 genomic copies per viral preparation). The numbers at the upper right corner denote the exposure times. stom, panc, int, sp, kid, br, liv, mscl: stomach, pancreas, intestine, spleen, kidney, brain, liver, muscle, respectively.
Extended Data Fig. 3 Genomic DNA and protein analyses of Lmna gRNA-treated mice.
a, Relative intensities of lamin A, progerin and lamin C bands on the immunoblot shown in Fig. 1e. The intensity of each band was normalized by α-tubulin intensity and compared to the normalized mock-control. b, Immunoblot of adult liver lysate from g1&2-treated (5 × 1011 genomic copies per viral preparation) heterozygous HGPS (Pro/+) with hemi- or homozygous Cas9 background showing that a single copy of Cas9 is sufficient. c–e, Deep sequencing was performed on liver and heart DNA to measure the level of in vivo indel formation by each gRNA. c, Scheme of the deep sequencing strategy. gLmna-1 and gLmna-2 target sites are denoted by red arrows. Ұ denotes HGPS mutation. PCR amplicons of deep sequencing are indicated by black curved arrows. The locations of the primers (Lmna F and Lmna R) used for the first round of nested-PCR are denoted by green arrows. Liver (d) and heart (e) DNA samples were collected from 2.5-month-old Cas9 mice that had been injected at P0 with either of the gRNA viruses (gLmna-1 or gLmna-2;1.5 × 1011 genomic copies per viral preparation), or with no virus. Treatment with one gRNA resulted in the presence of indels at their corresponding target sites rather than at the target site of the other gRNA. No indels were detected in the ‘no injection’ controls. Each treatment was evaluated in two independent in vivo experiments.
Extended Data Fig. 4 Physiological analyses of Lmna gRNA-treated mice.
a, Histological analysis of the aortic arch of untreated and gRNA-treated (g1&2) HGPS-Cas9 mice, 19 weeks old, injected at P0 with a 1:1 mix of gRNA1 and gRNA2 viral preparations. The black bracket shows the medial layer of the aortic arch. Scale bar, 100 μm. The bar graph on the right shows the number of nuclei in the medial layer (n = 21 for each group). b, Electrocardiographic analysis of untreated and gRNA-treated (g1&2) HGPS-Cas9 mice (untreated: n = 8; g1&2: n = 9). Heart rate is shown as beats per minute (bpm). c, Body weights of mice used for the grip strength test. Males and females are on the left and right, respectively. Cyan, red and green bars denote WT, negative control and treated mice, respectively. P values were calculated by unpaired Student’s t-test. Data are presented as mean ± s.e.m. In each box plot, the box extends from the 25th to 75th percentiles and the black lines in the middle represent the median. The whiskers show the upper and lower limits of the range; significance was determined by one-way ANOVA with Bonferroni correction.
Extended Data Fig. 5 Observed phenotypes of HGPS mice with extended lifespan.
a, Prevalence of megacolon/megacecum among homozygous HGPS mice treated with gLmna-1 and 2 (g1&2) versus negative controls. Asymp: asymptomatic (no obvious gastrointestinal phenotype); G.I: gastrointestinal phenotype characterized by megacolon/megacecum and inability to defecate. b, Chenges in body weight of a representative g1&2-treated homozygous mouse that died suddenly after 28 weeks. c, Necropsy of a g1&2-treated homozygous mouse exhibiting megacolon/megacecum (left and bottom) versus negative control (right).
Supplementary information
Supplementary Information
Supplementary Table 1
Supplementary Video 1
g1&2-treated homozygous mouse (active chinchilla colored) versus its heterozygous sibling (black) and a mock-treated homozygous mouse (inactive chinchilla colored). The treated and heterozygous mice were 19.4 weeks old, 3 weeks older than the mock-treated control. Same animals as in Fig. 1f.
Supplementary Video 2
Same animals as in Video 1, except the black mouse is the mock-treated WT sibling (same as in Fig. 1f) of the negative control.
Source data
Source Data Fig. 1
Source Data (Full Blots)
Rights and permissions
About this article
Cite this article
Beyret, E., Liao, HK., Yamamoto, M. et al. Single-dose CRISPR–Cas9 therapy extends lifespan of mice with Hutchinson–Gilford progeria syndrome. Nat Med 25, 419–422 (2019). https://doi.org/10.1038/s41591-019-0343-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41591-019-0343-4
This article is cited by
-
Methionine adenosyltransferase2A inhibition restores metabolism to improve regenerative capacity and strength of aged skeletal muscle
Nature Communications (2023)
-
Unique progerin C-terminal peptide ameliorates Hutchinson–Gilford progeria syndrome phenotype by rescuing BUBR1
Nature Aging (2023)
-
Progeroide Syndrome
Die Dermatologie (2023)
-
Transient expression of an adenine base editor corrects the Hutchinson-Gilford progeria syndrome mutation and improves the skin phenotype in mice
Nature Communications (2022)
-
SerpinE1 drives a cell-autonomous pathogenic signaling in Hutchinson–Gilford progeria syndrome
Cell Death & Disease (2022)